THE ARBELY LAB
RESEARCH
1
Lysine acetylation in the regulation of metabolic pathways
Thousands of lysine acetylation sites were recently discovered on non-histone proteins, suggesting that acetylation is a widespread and evolutionarily conserved post translational modification. Specifically, it has been found that most of the enzymes of metabolic processes are acetylated, implying that acetylation is key regulator of cellular metabolism. Our lab is interested in deciphring the role of specific lysine acetylation events in the regulation of central metabolic pathways, such as glycolysis and the pentose phosphate pathway. As misregulation of metabolic pathways is common to most transformed cells, we wish to understand the role of acetylation in metabolic reprogramming of cancer cells.
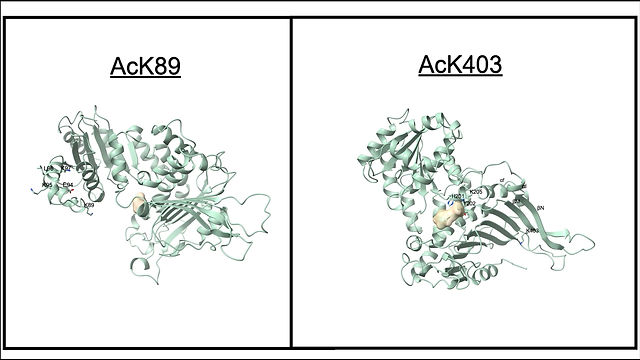
The movie illustrates the conformational change induced by K89 (left) or K403 (right) G6PD acetylation. Copyright © 2023 Fang Wu, Natali H. Muskat, Inbar Dvilansky, Omri Koren, Anat Shahar, Roi Gazit, Natalie Elia & Eyal Arbely
2
Lysine de-acetylation
One of the most pressing questions in the field of lysine acetylation is how lysine de-acetylases recognize and differentiate between their different substrates. Current methods for studying de-acetylation reactions are usually based on in vitro measurements with acetylated peptides. Our aim is to unravel the molecular basis of deacetylase-substrate specificity by studying the deacetylation of full-length acetylated proteins. In addition, we develop new technologies and methodologies to facilitate structure-function studies of deacetylase and acylated substrates.
3
Genetic code expansion technology
The translation machinery has been evolved over hundreds of millions of years to efficiently translate triple-nucleotide codons, each encoding one of the canonical amino acids. Yet, this machinery is far from exploiting its full potential. For example, one of the 64 triple-nucleotide codons can be reassigned to encode the site-specific and co-translational incorporation of a non-canonical amino acid into proteins, thereby creating a genetic code composed of more than 20 acids. The experimental approach (termed genetic code expansion), enables the introduction of distinct chemical moieties into full-length proteins expressed in living organisms, and was pioneered by Prof. Peter G. Schultz (Scripps Research Institute). Our lab is interested in further developing this approach to enable robust and biologically relevant studies in cultured mammalian cells.